Calculate molecular descriptors and fingerprints from SMILES and store them in a data frame [Python, RDKit]
How to create a data frame containing molecular descriptors and fingerprints from SMILES of a compound dataset with RDKit. Even if I tried to make my own QSAR / machine learning model, I stumbled upon creating molecular descriptors and fingerprints, so I will summarize them below.
Significance of storing in data frame
You can create a list type just to put it in machine learning, but using a data frame makes it easier to do the following.
- A bird's-eye view of the compound dataset based on the descriptor / fingerprint created
- Data preprocessing such as missing value handling and dimension reduction
Also, in RDKit, SMILES is once converted to a mol object in order to calculate the descriptor, but even if there is something that could not be converted well at that time, the data frame is easier to handle.
Try to practice
Preparation
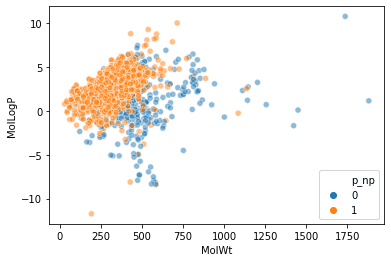
For sample data, we will use SMIELS from MoleculeNet's BBBP (blood-brain barrier penetration dataset).
The RDKit mol object is stored in a column called ROMol, so create a descriptor based on this.
Reference: List of compound datasets
import numpy as np
import pandas as pd
from rdkit import rdBase, Chem
from rdkit.Chem import AllChem, PandasTools, Descriptors
from rdkit.Chem.Draw import IPythonConsole
print('rdkit version: ',rdBase.rdkitVersion) # rdkit version: 2019.03.4
# 下準備
# データセットの読み込み
df = pd.read_csv("BBBP.csv")
# dfのSMILES列を参照してMolオブジェクト列をデータフレームに加える
PandasTools.AddMoleculeColumnToFrame(df,'smiles')
# Molオブジェクトが作成できたか確認
print(df.shape)
print(df.isnull().sum()) (2050, 4) num 0 name 0 p_np 0 smiles 0 ROMol 11 dtype: int64
The error "Explicit valence for atom # 1 N, 4, is greater than permitted" is displayed because there was a molecule (ion, etc.) with an abnormal valence ("4 atoms in N"). The value is above the permissible value. ")None was returned to the ROMol of such a molecule, and there were 11 such SMILES here.
You can deal with them one by one, but if the number is small, it is quick to remove them for the time being.Therefore, use isnull (). Sum () to check for missing values in the ROMol column, and remove those rows if any.
Reference: Troubleshooting compound data reading
# ROMolが作成できなかったものを確認
print(df[df.ROMol.isnull()])
# 欠損行の除去
df = df.dropna() 
If you see "WARNING: not removing hydrogen atoms without neighbors", it is probably because the data contains salts. RDKit saves H by default, so if there is an H (salt, etc.) that is not bound to the neighbor, such H cannot be removed and warns you.
Creating a molecular descriptor
The Map function is useful for applying a function to an object in each row in a data frame.
Since the descriptor names and functions are listed in "Descriptors.descList" of RDKit, it takes a little time, but I was able to calculate all at once with the for function and map function and return it to the data frame.
for i,j in Descriptors.descList:
df[i] = df.ROMol.map(j)
df.shape
# (2039, 205)
df.head()
Descriptors for 201 columns have been added.
If you apply the resulting variables to scikit-learn or a deep learning framework, you may get the error "Value Error: Input contains NaN, infinity or a value too large for dtype ('float64')". Was fine if I did the following.
for i,j in Descriptors.descList:
df[i] = df['ROMol'].map(j)
df['Ipc'] = [Descriptors.Ipc(mol, avg=True) for mol in df['ROMol']] It seems that the cause is that a part of the descriptor "IPC" value is created as large as infinite.
Reference: # 12 What to do if the IPC value of the RDKit 2D descriptor is very large
Reference: List of molecular descriptors
Creating a fingerprint
It can be calculated quickly using the apply function, but it seems that the list of fingerprints is stored in one column.
Since the fingerprint is saved in the format of Explicit BitVect object, it took some time to store each value in one column.
# 下準備
df = pd.read_csv("BBBP.csv")
PandasTools.AddMoleculeColumnToFrame(df,'smiles')
df = df.dropna()
# 1列にfingerprintのリストを追加する場合
df['FP'] = df.apply(lambda x: AllChem.GetMorganFingerprintAsBitVect(x.ROMol, 2, 1024), axis=1)
# fingerprintの各値を各列に格納する場合
# 個別に01をデータフレームに格納する
FP = [AllChem.GetMorganFingerprintAsBitVect(mol, 2, 1024) for mol in df.ROMol]
df_FP = pd.DataFrame(np.array(FP))
# フィンガープリントをもとのデータフレームに結合
df_FP.index = df.index
df = pd.concat([df, df_FP], axis=1)









In-Depth Discussions
Comment list
There are not any comments yet